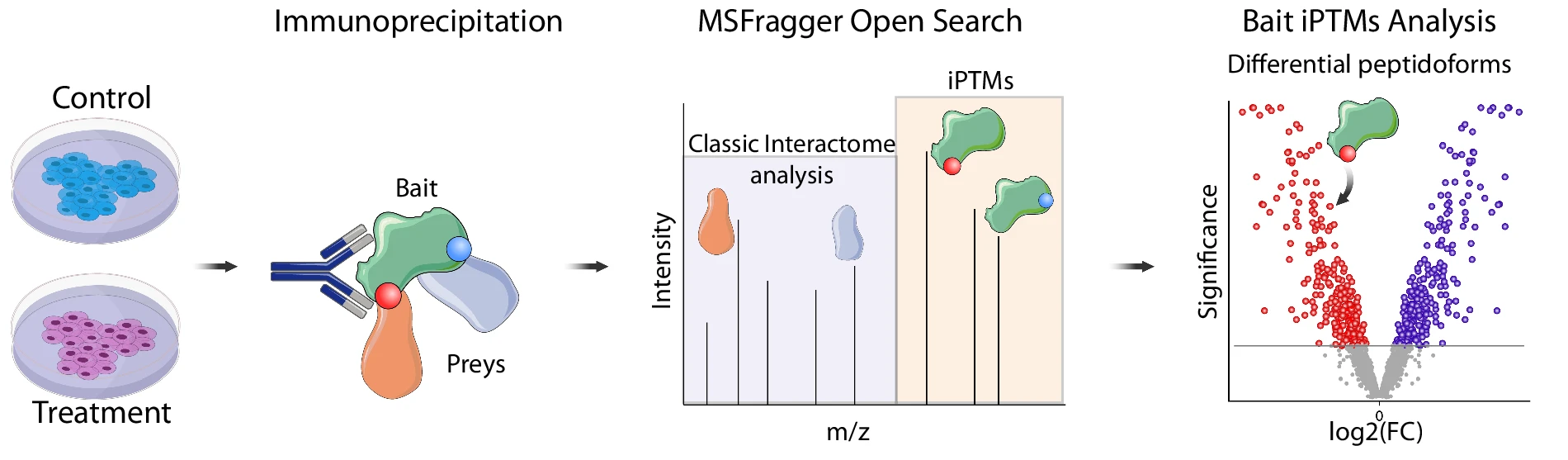



In an exciting new strategy, Savvas hypothetized that antibody enrichment would enrich modified (PTMs) and mutated bait peptides. Using MSFragger Open Search (Nesvilab) coupled to SAINTexpress, we identify differentially present peptidoforms.
By re-analyzing the publicly available #HRAS WT vs. G12D interactomes, we specifically identify the positive control LVVVGAGGVGK peptidoforms in its mutated and WT state among the most significant hits of our analysis.
Finally, to test our metholodogy against an endogenously expressed bait, we reanalysed #BRD4 IP-MS in 4 leukemia cell lines. Specific phosphorylated peptides were differentially present between these leukemia cell lines suggesting different BRD4 proteoforms in unpertubed cells.
A big thank you to @CRGenomica, @ERC_Research for supporting our work. And a huge Congrats to @Kourtissavvas and the rest of the authors for this intuitive work that will reshape the way we look at IP-MS data!
Read the original publication at Nature Scientific Data
| Cookie | Duration | Description |
|---|---|---|
| cookielawinfo-checbox-analytics | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Analytics". |
| cookielawinfo-checbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checbox-others | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Other. |
| cookielawinfo-checkbox-advertisement | 1 year | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Advertisement". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-performance | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Performance". |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
| Cookie | Duration | Description |
|---|---|---|
| _ga | 2 years | This cookie is installed by Google Analytics. The cookie is used to calculate visitor, session, campaign data and keep track of site usage for the site's analytics report. The cookies store information anonymously and assign a randomly generated number to identify unique visitors. |
| Cookie | Duration | Description |
|---|---|---|
| _ga_F3JJLHN009 | 2 years | No description |

