
Write us:
info_sdelcilab@crg.eu

The central role of metabolic rewiring during cancer progression is undeniable, but its direct impact on chromatin functions has been poorly investigated.
We recently described that enzymes of central metabolism can localize on chromatin in cancer cells. However, their chromatin-associated roles remain mostly elusive. Therefore, the scope of our research is to dissect the role of enzymatic activities on chromatin, with a particular focus on cancer. To do so, we apply chemical biology and functional genomics approaches to unbiased chromatin reporters (REDS) for selected metabolic enzymes. We aim to identify the molecular networks defining the direct interplay between chromatin and cancer metabolism. Our research focuses on a brand-new area of cancer biology and has the potential to uncover novel chromatin-associated metabolic vulnerabilities.
The breakthrough aspect of our investigation has been recognized by the European Research Council which, in 2019, awarded us with one of the prestigious ERC starting grant.
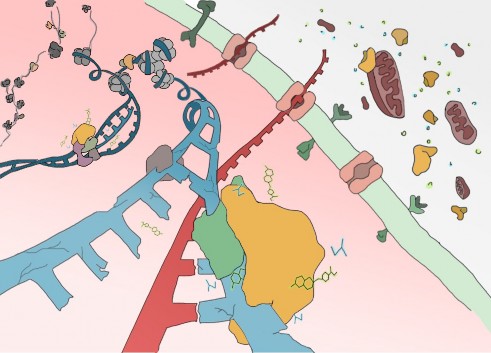
Epigenetic regulation and metabolism are of great interest in cancer research. However, physical and functional connections between these two areas remain largely unexplored. While it is commonly believed that metabolites can randomly distribute inside the cell, recent evidence rather favors the hypothesis that production of certain metabolites in specific subcellular compartments orchestrates different cellular processes.
EPICAMENTE aims at exploring whether the localization of enzymatic activities on chromatin can integrate cancer metabolism with chromatin remodeling to control epigenetic regulation and tumor progression.
First, I aim at providing a dataset of chromatin-bound metabolic enzymes in a comprehensive panel of cancer cell lines. By combining a chromatin fluorescent reporter cell line strategy with epigenomic approaches, I will define the epigenetic and transcriptional scenarios orchestrated by chromatin-bound metabolic enzymes, and investigate their relevance in cancer cell proliferation.
Performing genetic screenings with the chromatin fluorescent reporter cell lines will allow the identification of genetic interactors mediating the epigenetic role of chromatin-bound metabolic enzymes. In parallel, I aim to screen for small molecules able to counteract the epigenetic states mediated by those metabolic enzymes.
Finally, I will validate my results in in vivo cancer models, thus adding an important translational aspect to the project, and opening up new opportunities for cancer therapy.
In most cases, the belief is that intracellular materials reside inside steady-state membrane-based compartments, which limit the interactions between different molecular pathways. By describing the role of chromatin-bound metabolic enzymes and discovering direct connections between cancer metabolism and epigenetic regulation, I will scrutinize this belief.
| Cookie | Duration | Description |
|---|---|---|
| cookielawinfo-checbox-analytics | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Analytics". |
| cookielawinfo-checbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checbox-others | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Other. |
| cookielawinfo-checkbox-advertisement | 1 year | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Advertisement". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-performance | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Performance". |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
| Cookie | Duration | Description |
|---|---|---|
| _ga | 2 years | This cookie is installed by Google Analytics. The cookie is used to calculate visitor, session, campaign data and keep track of site usage for the site's analytics report. The cookies store information anonymously and assign a randomly generated number to identify unique visitors. |
| Cookie | Duration | Description |
|---|---|---|
| _ga_F3JJLHN009 | 2 years | No description |

